
Goals
CropXR develops smart data breeding methodology to construct integrated computer models of plants. The models demonstrate how plants respond to different stresses. This will help breeders to grow crops (which are cultivated plants grown for food) in an optimal way. These cutting-edge models are developed by collecting a large volume of measurements and combining these with various types of already available data and prior knowledge. The first models will be based on the plant Arabidopsis (thale cress). Subsequently, they will be deployed to six selected crops. Eventually, this technology should be available for all kinds of crops.

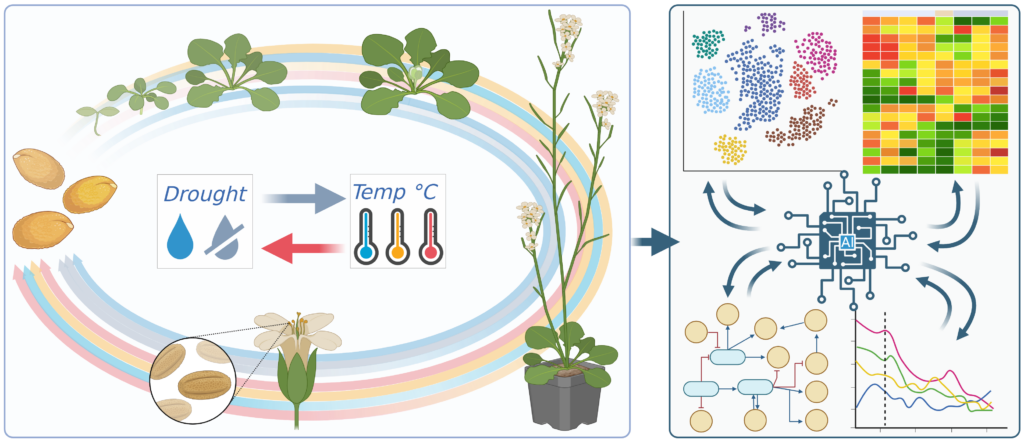
Approach
The Translator team works in an iterative manner. First, it translates the model for heat/drought stress response in Arabidopsis seedling establishment to three evolutionary closely related crops for which data is already available. Subsequently, the team translates the model to three other crops which are more challenging as their evolutionary relatedness is further removed from Arabidopsis. For each crop, the team develops methods to combine existing knowledge with new measurement data. This should lead to crop-specific integrated models that enable measurement. Moreover, the realization of these crop-specific models require less time and effort since they build on already existing insights. It is expected that after a few years, the developed methodology can support the translation of integrated models to traits and stresses relevant to breeding.
Activities
The team consists of experimental and computational biologists. The experimentalists start by performing seed establishment studies in a number of crops. This supports the translation of an earlier developed model. At the same time, the computational scientists set up methods to translate gene expression modules and mechanistic models between plants. This will lead to integrated models. The next step is to fine-tune the crop measurement data in an iterative manner. Eventually, the experimentalists will validate a few key findings in the lab. This will be the starting point for further activities that lead to actual application in crop breeding.
Team
Work package leader:
Dick de Ridder, Professor Bioinformatics, WUR
Annabel Dekker, Bioinformatician, ENZA – industrial partner
Basten Snoek, Assistant Professor Bioinformatics, UU
- Basten Snoek, Assistant Professor Bioinformatics, UU
- Berend Snel, Professor Bioinformatics, UU
- Guido van den Ackerveken, Professor Translational Plant Biology & Scientific Director CropXR, UU
- Janne Bibbe, PhD candidate, UU
- Jochem Evers, Professor Crop Physiology, WUR
- Kimm van Hulzen, Head of Genetics and Breeding Group, Genetwister – industrial partner
- Kirsten ten Tusscher, Professor Computational Developmental Biology, UU
- Lidija Berke, Group Leader Bioinformatics and Software Development, Genetwister – industrial partner
- Luca Laurenti, Assistant Professor Systems & Control, TU Delft
- Manuel Mazo Espinosa, Associate Professor Systems & Control, TU Delft
- Marcel van Verk, Team Leader Crop Data Science, Keygene – industrial partner
- Marco Busscher, Technician, WUR
- Marrit Alderkamp, Technician, UU
- Monica Garcia Gomez, Assistant Professor, UU
- Nikkie van Bers, Head of Innovation and Application, Genetwister – industrial partner
- Petra Bleeker, Associate Professor Plant Physiology and Signal Transduction, UvA
- Rashmi Sasidharan, Professor Plant Stress Resilience, UU
- Richard Immink, Professor Plant Reproduction, WUR
- Ronald Pierik, Professor Molecular Biology, WUR
- Xenja Ploeger, Technician, WUR

